BioMedGPS Data¶
A repo for building a knowledge graph and training knowledge graph embedding models for drug repurposing and disease mechanism research.
Follow the DocWebsite to learn more about this project. This DocWebsite is built on all markdown files in this repository. So you can also read the markdown files in this repository to learn more about this project.
Introduction¶
A knowledge graph is a graph-structured database that contains entities and relations. The entities are the nodes in the graph and the relations are the edges in the graph. The knowledge graph can be used to represent the biomedical knowledge and the relations between entities. A biomedical knowledge graph can be used for drug repurposing and disease mechanism research. Such as:
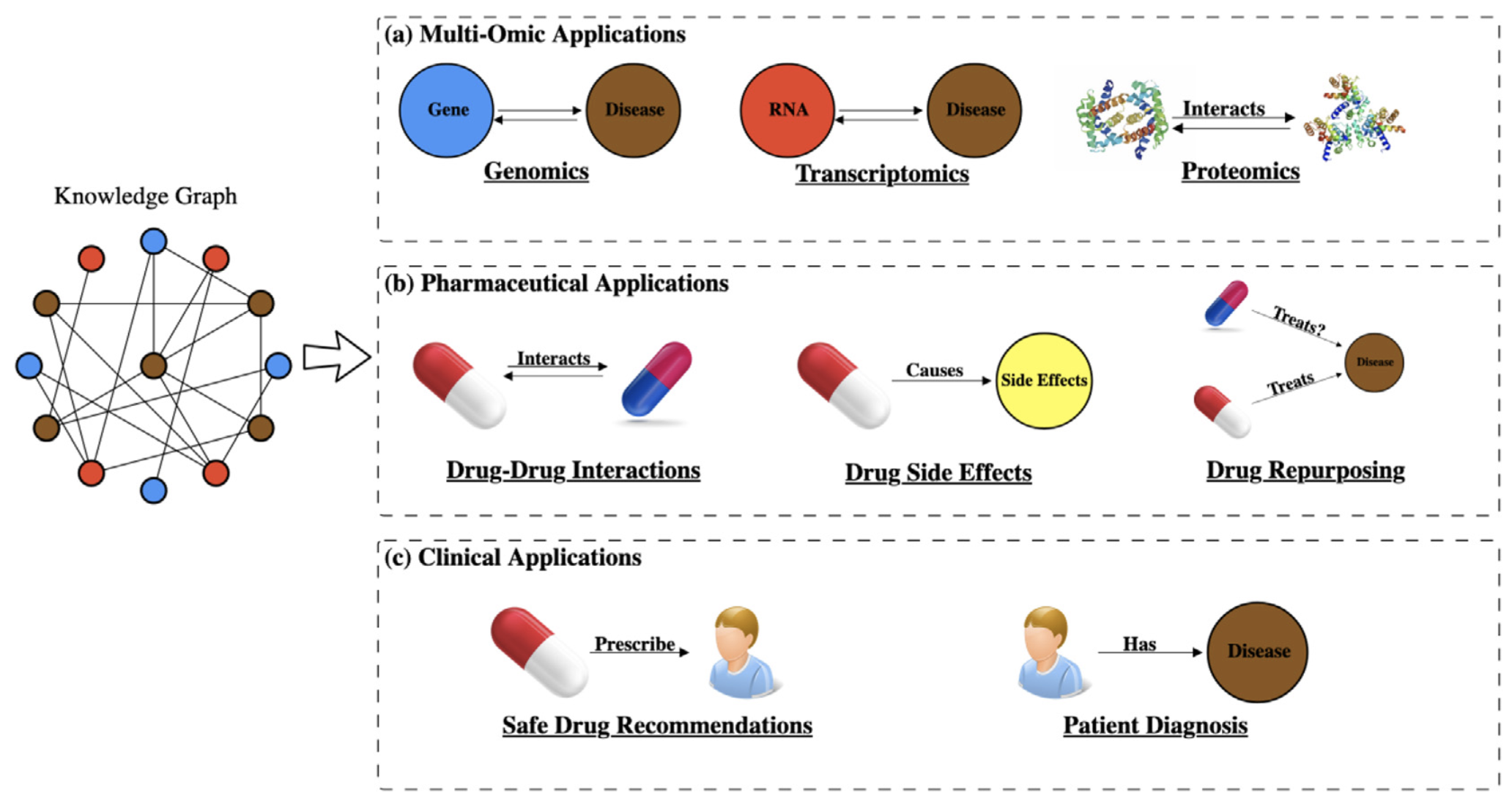
From Nicholson et al. CSBJ 2020.
The knowledge graph can be used to train knowledge graph embedding models.
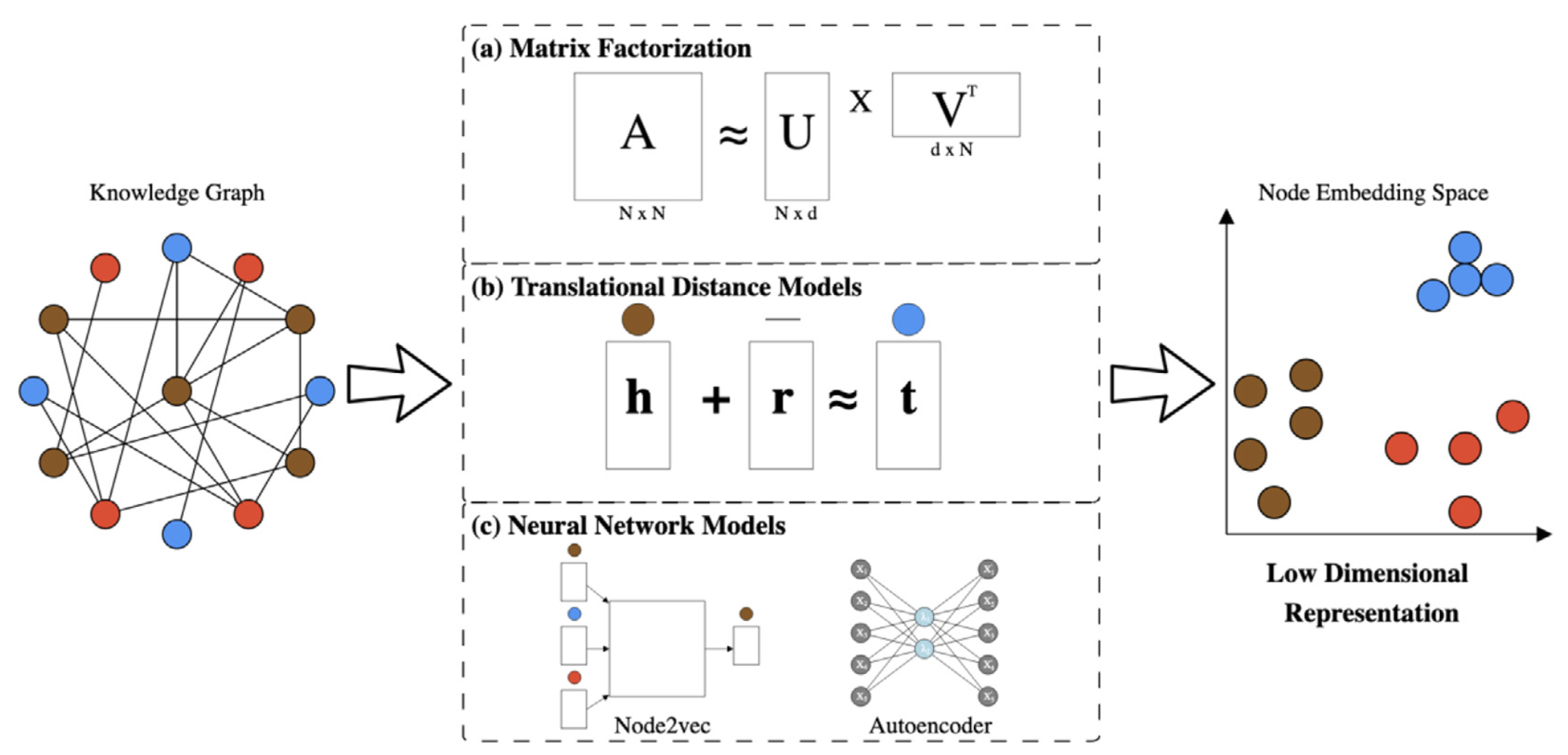
From Nicholson et al. CSBJ 2020.
But before that, we need to do some preprocessing to build a knowledge graph. Such as Entity Alignment, Entity Disambiguation. The following figure shows the key steps in the project.
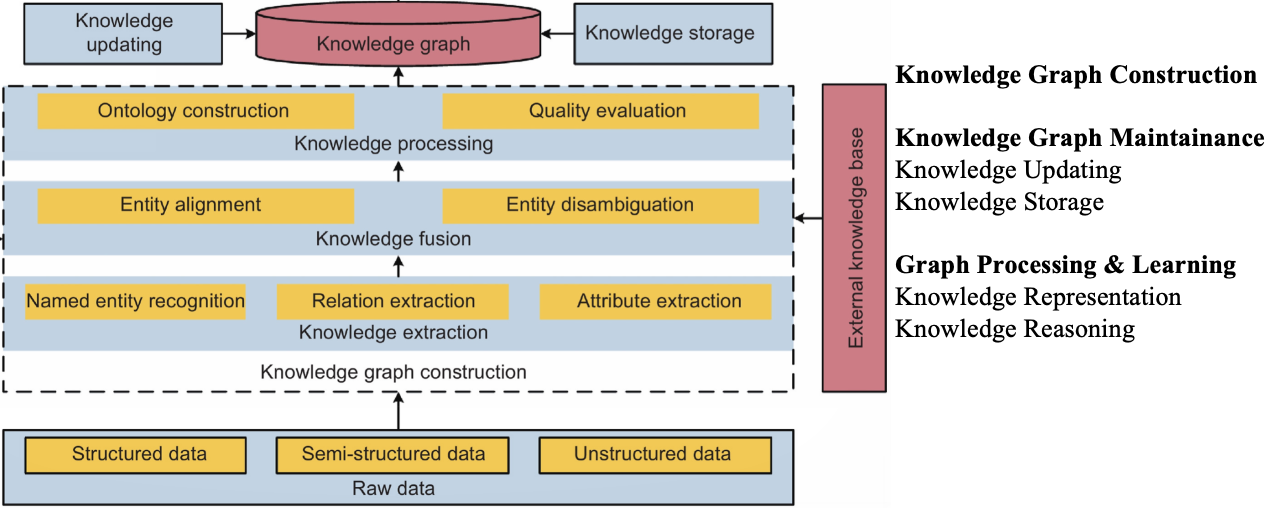
Unknown Source [TBD]
Before you start, I recommend you to read the following papers:
-
Barabási, A.-L., Gulbahce, N. & Loscalzo, J. Network medicine: a network-based approach to human disease. Nat. Rev. Genet. 12, 56–68 (2011).
-
Nicholson, David N., and Casey S. Greene. "Constructing knowledge graphs and their biomedical applications." Computational and structural biotechnology journal 18 (2020): 1414-1428.
-
Ioannidis, Vassilis N. and Song, Xiang and Manchanda, Saurav and Li, Mufei and Pan, Xiaoqin and Zheng, Da and Ning, Xia and Zeng, Xiangxiang and Karypis, George. DRKG - Drug Repurposing Knowledge Graph for Covid-19. PDF
Key Steps in the Project¶
If you want to use the pre-built knowledge graph and the pre-trained knowledge graph embedding models, you can skip the following steps and access our online service.
If you only want to use and analyze the pre-built knowledge graph, you can follow the instructions in the README.md file to download the pre-built knowledge graph. After that, you can see the graph_analysis directory to analyze the knowledge graph.
If you are interested in how the training scripts work, you can see the examples directory in this repository.
Please note that it is not necessary to run all the following steps in the project. You can run the steps you are interested in. But you need to make sure the dependencies among the steps. For example, if you want to train the knowledge graph embedding models, you need to build a knowledge graph or download the pre-built knowledge graph first. If you want to analyze the knowledge graph embedding models, you need to train the knowledge graph embedding models first.
Step 1: Install dependencies¶
More details can be found in the Install Dependencies file.
Step 2: Build & Analyze a knowledge graph¶
This repository contains the codes to build a knowledge graph for BioMedGPS project. Which depends on the ontology-matcher package and graph-builder package.
If you want to run the following codes to build a knowledge graph for BioMedGPS project, you need to install all dependencies first. Please see the Install Dependencies file.
After that, you can run the following codes to build a knowledge graph for BioMedGPS project.
NOTE: Be sure to activate the python environment you created and located in the root directory of this repository when running the following codes.
# Remove the following directories for a clean build
rm -rf ./graph_data/extracted_entities ./graph_data/formatted_entities ./graph_data/formatted_relations
python run_markdown.py ./graph_data/KG_README.md --run-all
# The run_markdown.py is a script to run the codes in a markdown file.
# It will extract the code blocks from the markdown file and run them one by one.
# If you want to run a specific code block, you can use the following command.
# If you see 'Cannot identify the language' message, this means that the code block is not necessary to run.
python run_markdown.py ./graph_data/KG_README.md
If you want to build a knowledge graph for BioMedGPS project step by step by yourself, you can follow the instructions in the KG_README.md file.
How to analyze the knowledge graph? More details can be found in the graph_analysis directory in this repository or see the related documentation graph_analysis/README.md.
Step 3: Train, Evaluate, Analyze & Benchmark KGE models¶
-
Train & evaluate knowledge graph embedding models
If you want to train the knowledge graph embedding models by yourself, you can see the wandb directory in this repository or see the related documentation wandb/README.md.
-
Benchmark knowledge graph embedding models
If you want to benchmark the knowledge graph embedding models, you can see the benchmarks directory in this repository or see the related documentation benchmarks/README.md.
-
Analyze the knowledge graph embedding models
If you want to analyze the knowledge graph embedding models, you can see the embedding_analysis directory in this repository or see the related documentation embedding_analysis/README.md.
Step 4: Link Prediction¶
If you want to predict the relations between entities, you can see the prediction directory in this repository or see the related documentation prediction/README.md.
Step 5: Explain the prediction results¶
More details can be found in the biomedgps-explainer repository.